An interesting example of a new knotted fold is provided by the proteins with a novel 83 knot type. We found more than 250 such proteins in various bacteria (only in this type of organism), with high structural similarities. All of them are all-alpha proteins composed of (about 15) α-helices. As common for other folds, these proteins share structural features despite being sequentially distinct (as low as 10% sequence identity). Not only the 83 knot was never before seen in a protein, but also the fold is unique since none of the proteins has a homolog with an experimentally verified structure. If the knotting and the fold are predicted correctly, it would show that the Machine Learning methods are capable of modeling previously unknown and topologically complex proteins.
The length of the proteins varies from 418 to 560 amino acids, with the knot encompassing most of the structure (tail varies between 2 to 14 amino acids, e.g. in the case of A0A7X8EQ82 both knot tails possess 14 amino acids, see Fig. 1). In the example protein from Epulopiscium sp. (UniProtKB ID: A0A7X8EQ82) the knot is positioned between the 23 and 457 residue, leaving 22 and 21 amino acid long N- and C-tails, respectively (Figure 1A). This is a high-quality region of the model with an average pLDDT equal 90.4.
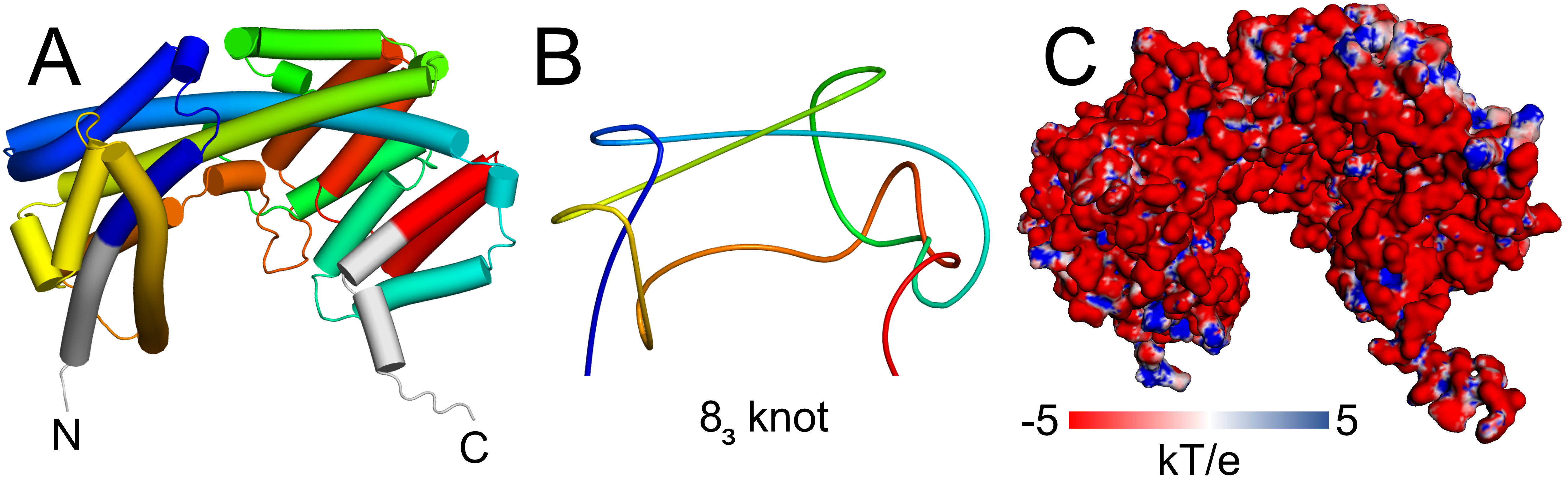
Figure 1. Example of a novel 8_3 knotted protein (UniProtKB ID: A0A7X8EQ82). A. Cartoon representation of the full-length structure with the knot shown in rainbow coloring and its tails in gray. B. Simplified scheme showing 8_3 knot generated with knot_pull [5]. C. Surface representation colored by an electrostatic potential calculated with APBS 3.4.1 in pH 7.
The protein has a high number of charged residues, constituting about 29% of the residues. Thus, the protein has a negative net charge (-30 in pH 7) that forms a vast negatively charged surface (Figure 1B). It is possible that due to this charge and the shape of the protein, it functions in the cell in a bound form, e.g. in a complex with a positively charged protein.
Within the AlphaFold predicted models we found proteins that show another take on the known, however unpopular, motif - β-helices. They are formed by twisting β-sheets around an axis to form a helical structure. Several different types of β-helices are already known due to the crystallization efforts, however, none of the structures have non-trivial topology. We found a group of over 180 bacterial proteins with a conserved trefoil (31 knot) motif in their C-terminal part. The proteins can have additional domains (mostly short transmembrane α-helices) besides the β-helical one which is quite small (around 130 amino acids). Based on the length and the way the β-helix is arranged (triangle-type cross-section), the knotted structures resemble insect antifreeze proteins (Figure 2).
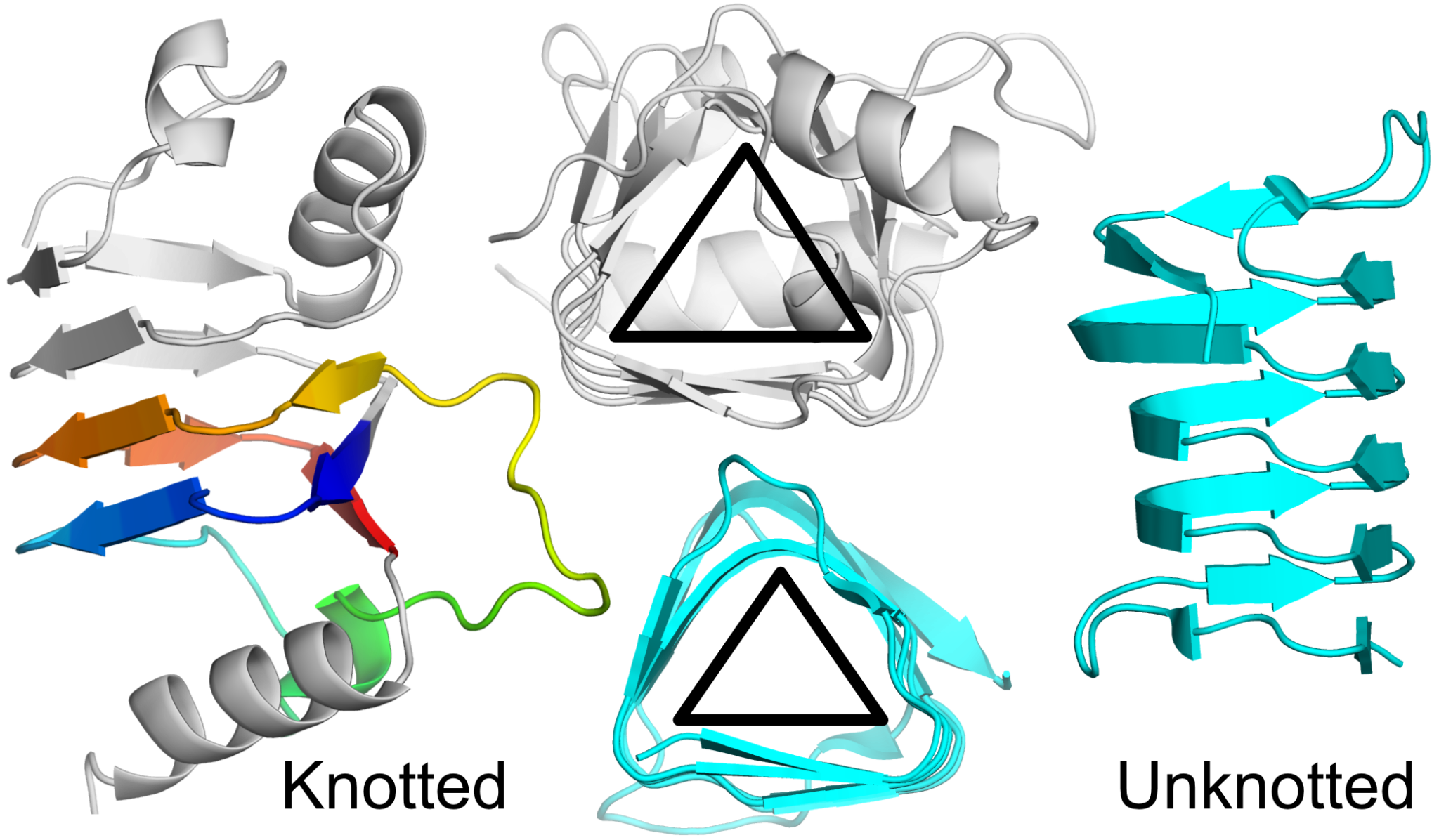
Figure 2. β-helical proteins with different topologies. Left side: 31 knotted model from Brevundimonas mediterranea (UniProtKB ID: A0A7W6A5G1) colored gray and the knot rainbow. Right side: unknotted antifreeze protein from Choristoneura fumiferana (Spruce budworm moth; PDB ID: 1l0s) colored cyan. Center: cross-sections of the structures.
The example knotted protein (UniProtKB ID: A0A7W6A5G1) is 127 amino acid long mainly beta protein. It has 10 β-sheets and 3 α-helices. The α-helices are capping the N- and C-terminal β-sheets to prevent aggregation. A similar security measure is found in other β-helical proteins, such as pectate lyases. This suggests that these proteins are not found in complexes but rather act on their own.
The trefoil knot in this protein is a compact motif (54 residues) comprised of 4 β-sheets and a short single α-helix. The knot is present between 56 and 110 residues and the region is modeled with high confidence (pLDDT of 94.7). Just before the knot, there are two strictly conserved amino acids - glycine and proline, that enable the sharp turn of the structure that results in the knot formation. Based on proteins with known X-ray structures, it is known that Gly and Pro are common in the case of knotted proteins [2].
The data deposited in AlphaKnot can be used not only to check if your protein of interest is knotted but also to find unknotted structures. For example, the superfamily of SPOUT proteins is the biggest group of knotted proteins, with many structures resolved experimentally. There is no single instance of unknotted protein within this group. However, we found several models predicted by AlphaFold that are annotated as SPOUT proteins (InterPro ID: IPR029026) that did not have a knot. We can verify this surprising result using topology data found in AlphaKnot (we performed a similar analysis in [1]).
Unknotted protein annotated as SPOUT is 165 amino acid long (UniProtKB ID: A0A497EM03), which is not uncommon for this superfamily. However, its closest knotted homolog has 201 residues (high sequence identity: 75% relative to the shorter sequence). Both sequential and structural alignments show that the difference in length is due to the absence of the C-terminal part of the unknotted protein (Figure 3). Since this fragment provides the crossing necessary for knot formation, the knot is absent in this model.
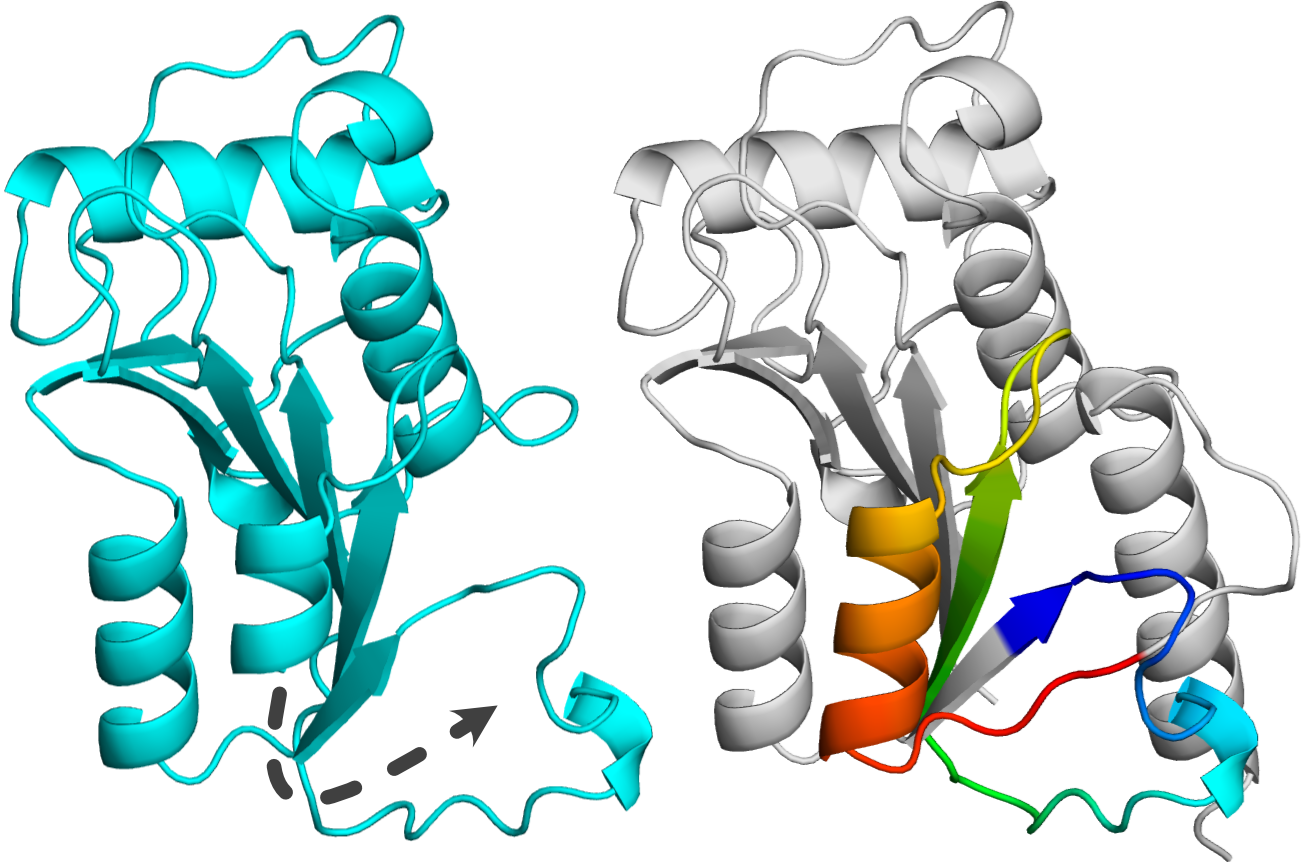
Figure 3. Homologous proteins from SPOUT superfamily. Left: unknotted protein (UniProtKB ID: A0A497EM03) with missing fragment shown in dashed line. Right: knotted protein (UniProtKB ID: A0A497F5R9) with the knot shown with rainbow coloring.
In the case of SPOUT proteins, the correctly folded knot is important for protein function as it creates part of the substrate binding site. Therefore, since the unknotted structure is missing this vital part of the sequence, it is representing a fragment of a protein. Overall, this means that the SPOUT superfamily does not have any truly unknotted proteins. Intersting similar situation can be observed in the case of other very well-known knotted families such as UCH [3] or membrane knotted proteins [4].